Taking New Aim at COVID-19
The coronavirus’s tangled strands of RNA could offer new ways to treat people who get infected
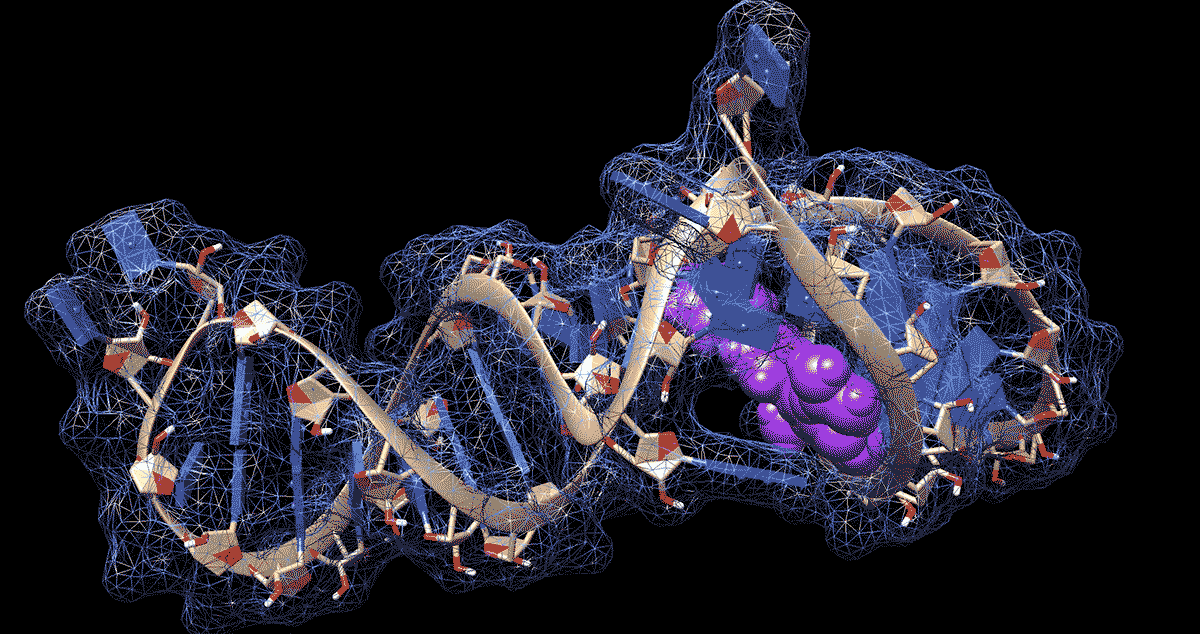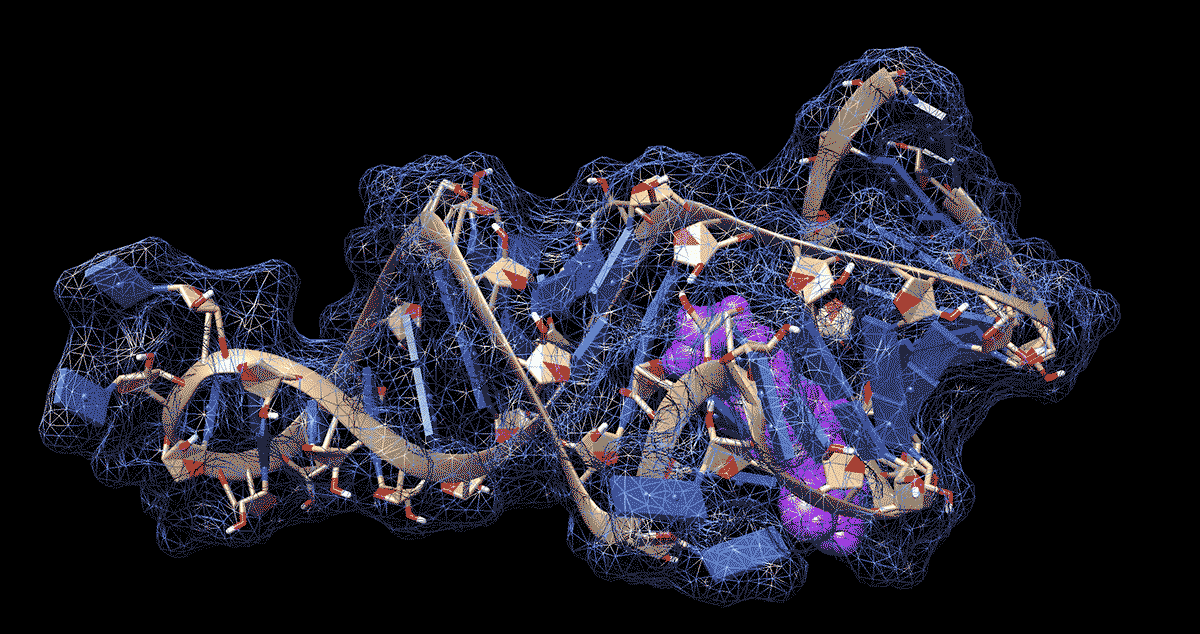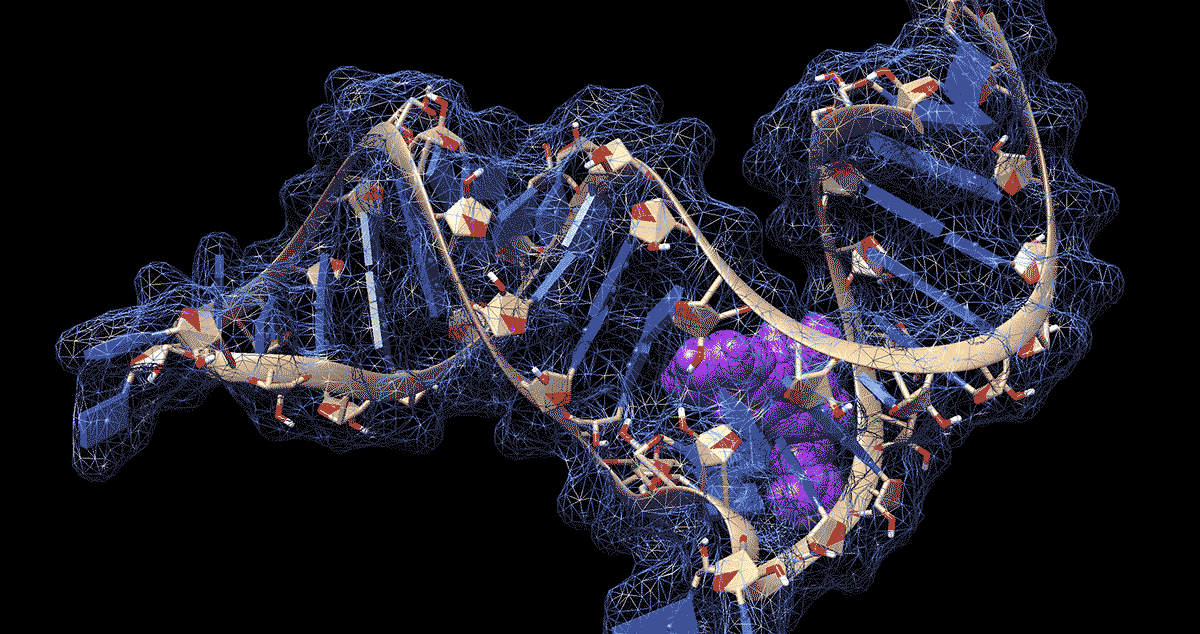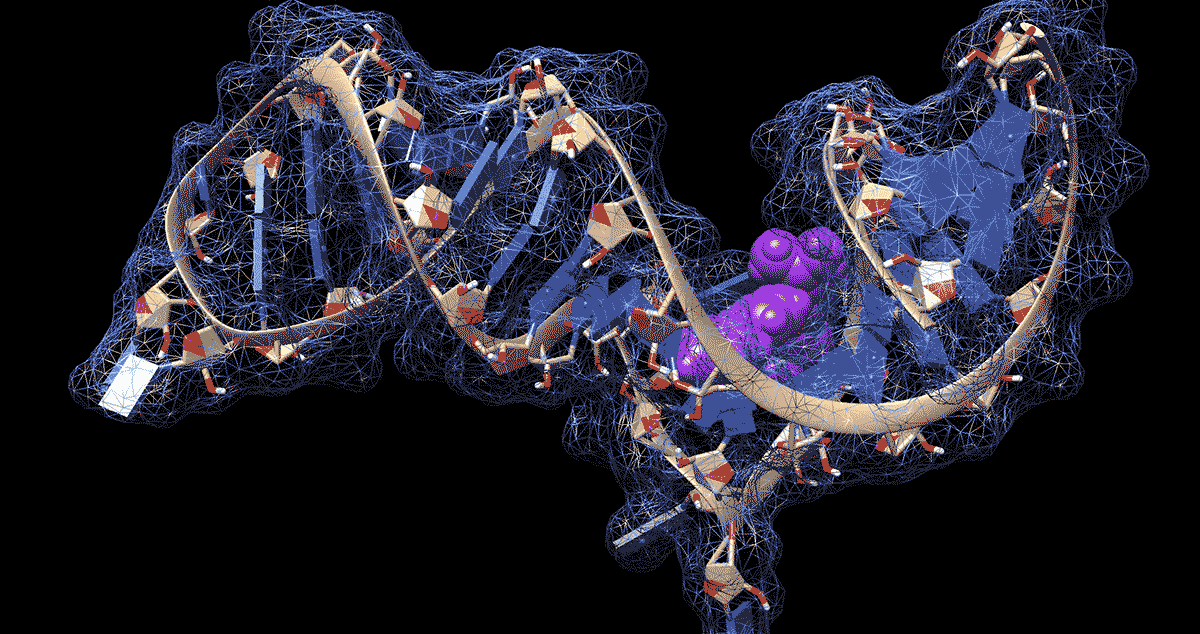
DURHAM, N.C. -- To the untrained eye, the loops, kinks and folds in the single strand of RNA that makes up the coronavirus genome look like a jumble of spaghetti or tangled yarn. But to researchers like Amanda Hargrove, a chemistry professor at Duke University, the complex shapes that RNA takes on as it folds upon itself could have untapped therapeutic potential in the fight against COVID-19.
 In a study published Nov. 26 in the journal Science Advances, Hargrove and colleagues have identified chemical compounds that can latch onto these 3D structures and block the virus’s ability to replicate.
In a study published Nov. 26 in the journal Science Advances, Hargrove and colleagues have identified chemical compounds that can latch onto these 3D structures and block the virus’s ability to replicate.
“These are the first molecules with antiviral activity that target the virus’s RNA specifically, so it's a totally new mechanism in that sense,” Hargrove said.
Even more than 18 months into the pandemic, that’s good news. We have vaccines to prevent COVID-19, but effective, easy-to-administer drugs to help people survive and recover once they’ve been infected remain limited.
The virus is receding in some parts of the world, but cases are still surging in others where vaccines are in short supply. And even in regions with easy access to vaccines, COVID-19 vaccine hesitancy means many of the world’s eight billion people remain vulnerable to infection.
To infect your cells, the coronavirus must break in, deliver its genetic instructions in the form of RNA, and hijack the body’s molecular machinery to build new copies of itself. The infected cell becomes a virus factory, reading the 30,000 nucleotide “letters” of the virus’s genetic code and churning out the proteins the virus needs to replicate and spread.
Most antivirals -- including remdesivir, molnupiravir and Paxlovid, the only antiviral drugs for COVID-19 that have been FDA-approved or are in line for approval -- work by binding to these proteins. But Hargrove and colleagues are taking a different approach. They’ve identified the first molecules that take aim at the viral genome itself -- and not just the linear sequence of A’s, C’s, G’s and U’s, but the complex three-dimensional structures the RNA strand folds into.
When the first terrifying hints of the pandemic started to make headlines, the team including Hargrove, Blanton Tolbert from Case Western Reserve University and Gary Brewer and Mei-Ling Li from Rutgers were already investigating potential drug candidates to fight another RNA virus -- Enterovirus 71, a common cause of hand, foot and mouth disease in children.
They had identified a class of small molecules called amilorides that can bind to hairpin-like folds in the virus’s genetic material and throw a wrench in the virus’s replication.
 To see if the same compounds could work against coronaviruses too, first they tested 23 amiloride-based molecules against another, far less deadly coronavirus responsible for many common colds. They identified three compounds that, when added to infected monkey cells, reduced the amount of virus within 24 hours of infection without causing collateral damage to their host cells. They also showed greater effects at higher doses. The researchers got similar results when they tested the molecules on cells infected with SARS-CoV-2, the virus that causes COVID-19.
To see if the same compounds could work against coronaviruses too, first they tested 23 amiloride-based molecules against another, far less deadly coronavirus responsible for many common colds. They identified three compounds that, when added to infected monkey cells, reduced the amount of virus within 24 hours of infection without causing collateral damage to their host cells. They also showed greater effects at higher doses. The researchers got similar results when they tested the molecules on cells infected with SARS-CoV-2, the virus that causes COVID-19.
Further work showed that the molecules stopped the virus from building up by binding to a site in the first 800 letters of the viral genome. Most of this stretch of RNA doesn’t code for proteins itself but drives their production.
The region folds in on itself to form multiple bulges and hairpin-like structures. Using computer modeling and a technique called nuclear magnetic resonance spectroscopy, the researchers were able to analyze these 3D RNA structures and pinpoint where the chemical compounds were binding.
The researchers are still trying to figure out exactly how these compounds stop the virus from multiplying, once they’re bound to its genome.
When it comes to using RNA as a drug target, Hargrove says the field is still in its early stages. Part of the reason is that RNA structures are unstable. They bounce around a lot more than their protein counterparts, which makes it hard to design molecules that can interact with them in specific ways
“The binding pocket that you’re looking for may not even be present most of the time,” Hargrove said.
What’s more, 85% of the RNA in an infected cell doesn’t belong to the virus, but to the ribosomes -- cellular particles made of RNA and protein -- of its human host. “There’s a sea of competition,” Hargrove said.
But Hargrove is hopeful. The first small-molecule drug that works by binding to non-ribosomal RNA directly, rather than proteins, was just approved by the FDA last August, to treat people with a devastating disease called spinal muscular atrophy. “So while there are lots of challenges, it’s not impossible,” Hargrove said.
The researchers have a patent pending on their method. They want to modify the compounds to make them more potent, and then test them in mice “to see if this could be a viable drug candidate,” Hargrove said.
This isn’t the first time that coronaviruses have caused an outbreak, and it likely won’t be the last, the researchers say. In the last two decades, the same family of viruses was responsible for SARS, which emerged in China and spread to more than two dozen countries in 2002, and MERS, first reported in Saudi Arabia in 2012.
The researchers determined that the loops and bulges of RNA they identified have remained essentially unchanged by evolution across related coronaviruses in bats, rats and humans, including the ones that caused the SARS and MERS outbreaks. That means their method might be able to fight more than just SARS-CoV-2, the virus that causes COVID-19.
Clearly, more antivirals would be valuable weapons to have, so when the next pandemic hits we’ll be better prepared. Having more drugs on hand would have another benefit: fighting resistance. Viruses mutate over time. Being able to combine drugs with different mechanisms of action would make it less likely that the virus could develop resistance to all of them simultaneously and become impossible to treat, Hargrove said.
“This is a new way to think about antivirals for RNA viruses,” Hargrove said.
The researchers collaborated across seven institutions for this study, including Rutgers University, Case Western Reserve University, Washington University School of Medicine in St. Louis, University of Nebraska-Lincoln, University of Glasgow and the University of Michigan.
This research was supported by the National Institute of General Medical Sciences (R35GM124785, GM126833), Tobacco Settlement Fund (21-5734-0010), Medical Research Council of the United Kingdom (MC_UU_12014/12), and Duke University.
CITATION: "Amilorides Inhibit SARS-CoV-2 Replication in vitro by Targeting RNA Structures," Martina Zafferani, Christina Haddad, Le Luo, Jesse Davila-Calderon, Liang Yuan-Chiu, Christian Shema Mugisha, Adeline Monaghan, Andrew Kennedy, Joseph Yesselman, Robert Gifford, Andrew Tai, Sebla Kutluay, Mei-Ling Li, Gary Brewer, Blanton Tolbert, Amanda Hargrove. Science Advances, Nov. 26, 2021. DOI: 10.1126/sciadv.abl6096