New Drug Candidate Found for Hand, Foot and Mouth Disease
Targeting RNA with small molecules could pave the way for new antivirals
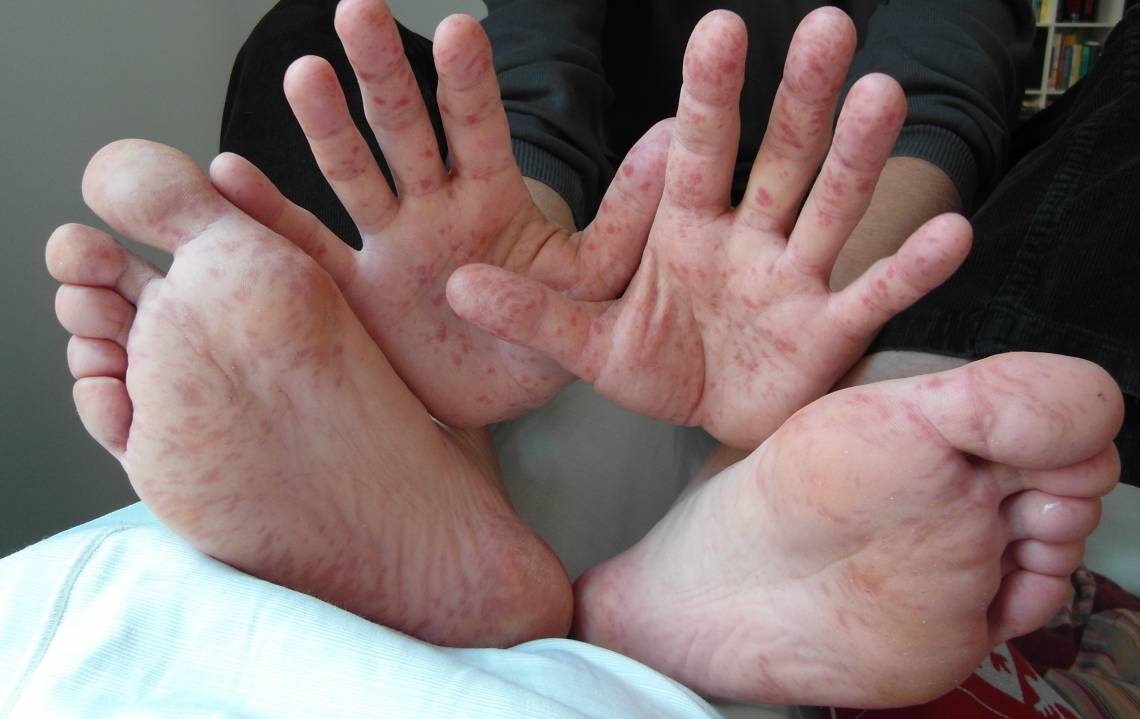
DURHAM, N.C. -- A study published this week in the journal Nature Communications offers some good news in the search for antiviral drugs for hard-to-treat diseases. Researchers have identified a potential new drug candidate against enterovirus 71, a common cause of hand, foot and mouth disease in infants and young children.
The compound of interest is a small molecule that binds to RNA, the virus’s genetic material, and changes its 3-D shape in a way that stops the virus from multiplying without harming its human host.
There are currently no FDA-approved drugs or vaccines for enterovirus 71, which affects hundreds of thousands of children each year, particularly in Southeast Asia. While most people get better within 7 to 10 days after suffering little more than a fever and rash, severe cases can cause brain inflammation, paralysis and even death.
The work could pave the way for new treatments for other viral infections as well, says a team of scientists at Duke University, Case Western Reserve University and Rutgers University.
Traditionally, most drugs are designed to bind to proteins to block or disrupt their role in causing disease. But much of the genome in humans and their microbial pathogens doesn’t code for proteins, which means that only a fraction of their genetic material is targeted by existing drugs.
“For diseases that don't have good treatments, maybe the problem is we've been targeting the wrong thing,” said co-author Amanda Hargrove, associate professor of chemistry at Duke.
Instead of targeting proteins, Hargrove and others are looking for small molecules that target RNA, which most drug discovery programs have overlooked.
When a virus like enterovirus 71 (or SARS-CoV-2, the virus that causes COVID-19) infects a human cell, it injects its RNA into the cell, hijacking the internal machinery to make copies of itself that eventually burst out to infect neighboring cells.
Previous work on enterovirus 71 singled out one part of its RNA structure that helps the virus co-opt the host machinery it needs to replicate. This RNA region folds over on itself to form a hairpin, with a bulge in the middle where unpaired nucleotides balloon out to one side.
If a drug can be developed to inhibit this region, the researchers say, we might be able to block the virus before it has a chance to spread.
For the current study, Hargrove and colleagues screened a library of some 30 small molecules, looking for ones that bind tightly to the bulge and not other sites in the virus’s RNA.
RNA is a wiggly molecule; when it binds to other molecules such as host proteins or small molecule drugs it takes on different 3-D shapes.
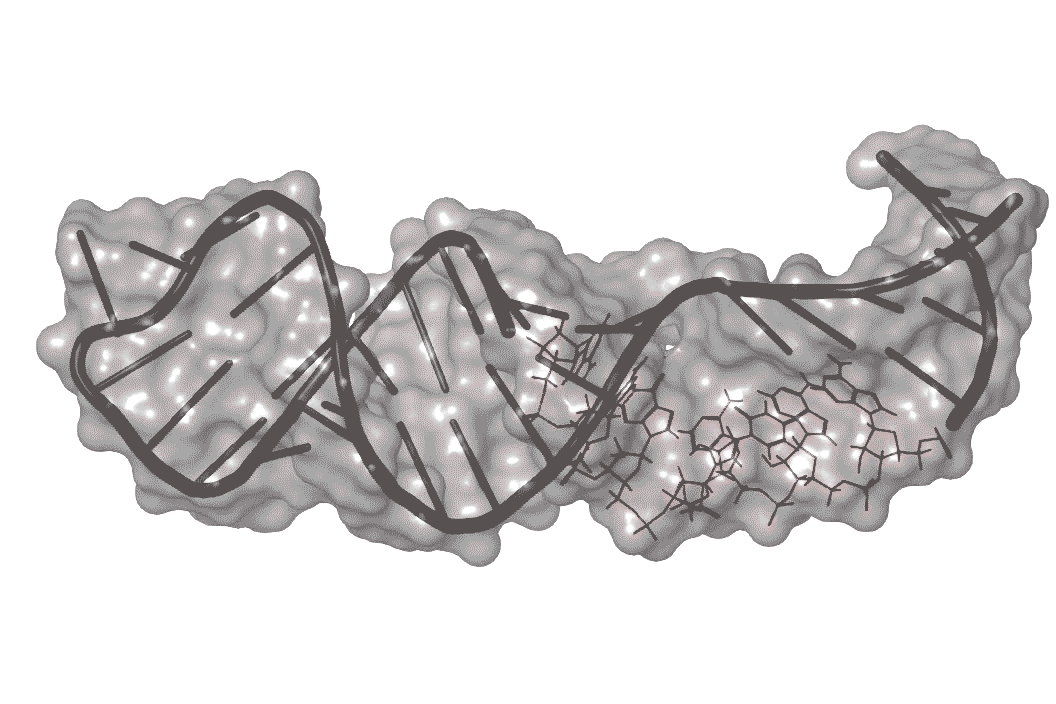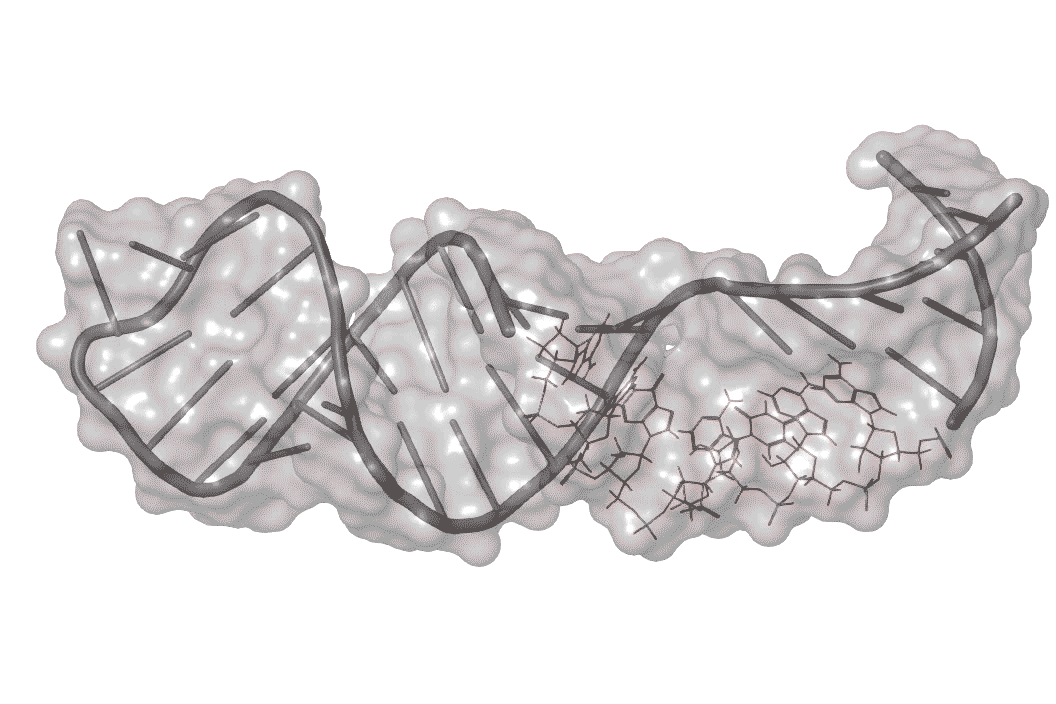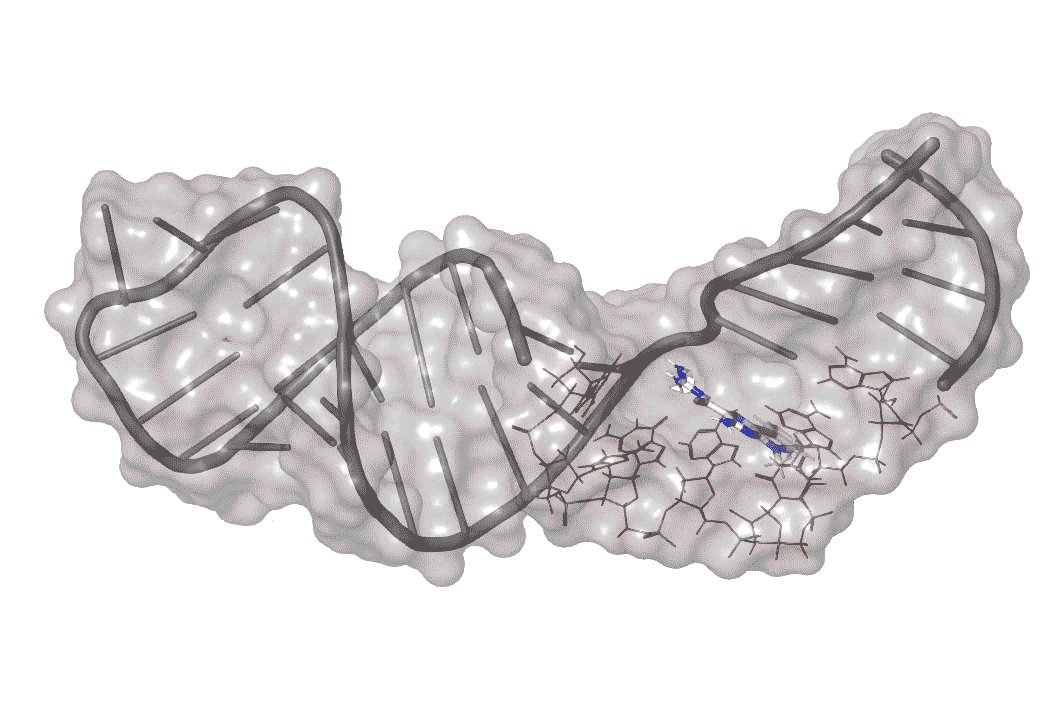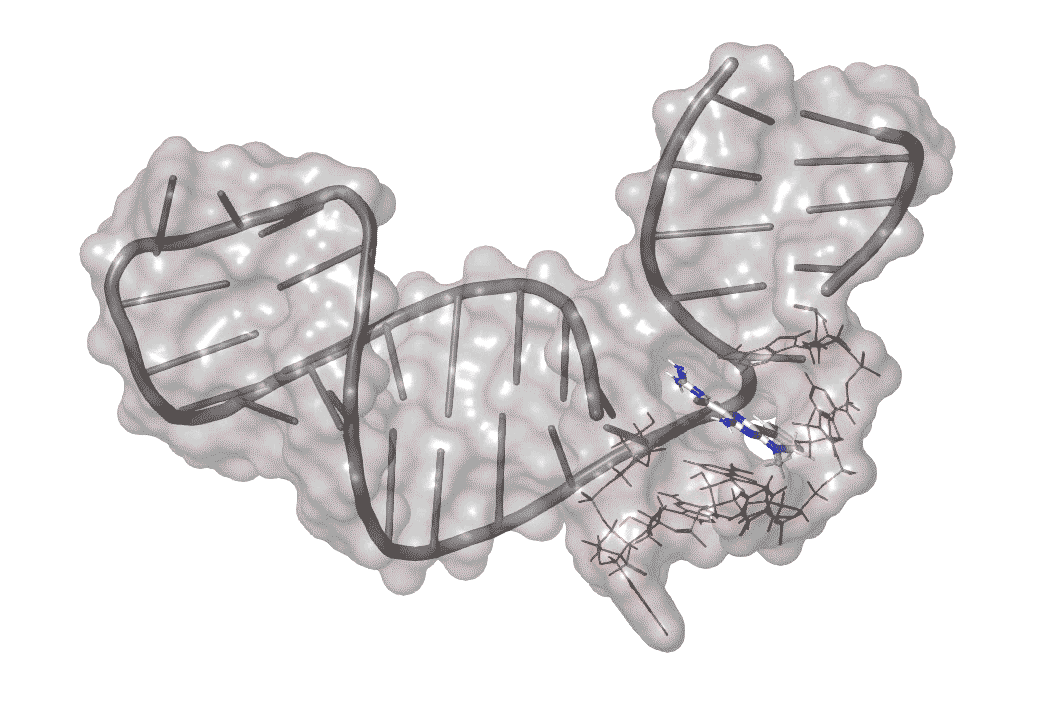
The researchers identified one molecule, dubbed DMA-135, that enters infected human cells and attaches itself to the surface of the bulge, creating a kink in this region.
This shape change, in turn, opens access to another molecule -- a human repressor protein that blocks the “reading out” of the virus’s genetic instructions, stopping viral growth in its tracks.
In an experiment, the researchers were able to use the molecule to stop the virus from building up inside human cell cultures in the lab, with bigger effects at higher doses.
Hargrove says it would take at least five years to move any new drug for hand, foot and mouth disease from the lab to medicine cabinets. Before their small molecule could reach patients, the next step is to make sure it’s safe and effective in mice.
In the meantime, the researchers are building on their success with enterovirus 71 and looking at whether RNA-targeting small molecules could be used to tackle other RNA viruses too, including SARS-CoV-2.
This research was supported by grants from the National Institutes of Health (U54 AI150470, R35 GM124785, R01 GM126833).
CITATION: "Small Molecule Targeting IRES Domain Inhibits Enterovirus 71 Replication via an Allosteric Mechanism that Stabilizes a Ternary Complex," Jesse Davila-Calderon, Neeraj Patwardhan, Liang-Yuan Chiu, Andrew Sugarman, Zhengguo Cai, Srinivasa R. Penutmutchu, Mei-Ling Li, Gary Brewer, Amanda E. Hargrove, Blanton S. Tolbert. Nature Communications, Sept. 22, 2020. DOI: 10.1038/s41467-020-18594-3