Duke Team Makes Cryo-EM Microscope Even Better Than Before
Software improvements speed data-acquisition by days
The amazing and Nobel prize-winning power of Cryo-EM microscopy to see the smallest details of molecules is about to grow greater.
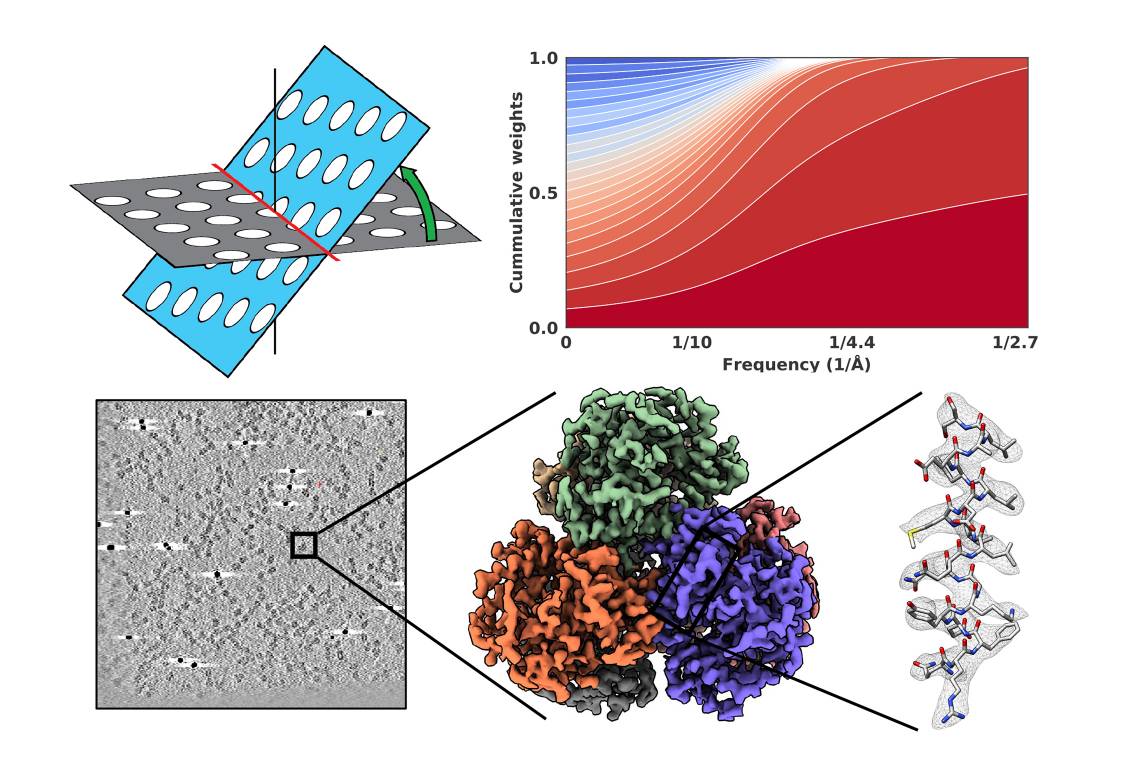
Duke researchers Alberto Bartesaghi, associate professor in Biochemistry and Computer Science and Mario Borgnia, Director of the Cryo-EM Core at the NIEHS in Research Triangle Park, published a paper this month in Nature Communications that describes scanning the smallest sample ever in the shortest time at the highest quality. Their new process also reduces data collection from ten days to one by speeding up the time it takes to render a 3D image.
Cryo-EM produces molecular-scale pictures by taking thousands of images and adding them together with software. This team improved on the process by accelerating the imaging of tilted specimens in a process called cryo-electron tomography, or cryoET.
Jonathan Bouvette, a postdoc in the Borgnia Lab, wrote the computer code that tells the microscope to take multiple images at different degrees of sample plate tilt, and Hsuan-Fu Liu, a PhD student in the Bartesaghi Lab, developed the computational algorithms to obtain the high-resolution structures.