DURHAM, N.C. -- Thumb through any old science textbook, and you’ll likely find RNA described as little more than a means to an end, a kind of molecular scratch paper used to construct the proteins encoded in DNA.
But over the last decade, scientists have begun to see RNA as an end in itself.
Research programs and biotech companies have sprung up with the mission of identifying small-molecule drugs that can target RNA to treat a variety of ailments like infectious diseases and muscular dystrophy. The trouble is, RNA is constantly bending, twisting and contorting its shape -- often within a few milliseconds. Researchers have found it hard to hit such a moving target.
“When it comes to targeting RNA, the devil is in the details, and the details are in the dynamics," said Hashim M. Al-Hashimi, Ph.D., James B. Duke Professor of Biochemistry and Chemistry at the Duke University School of Medicine.
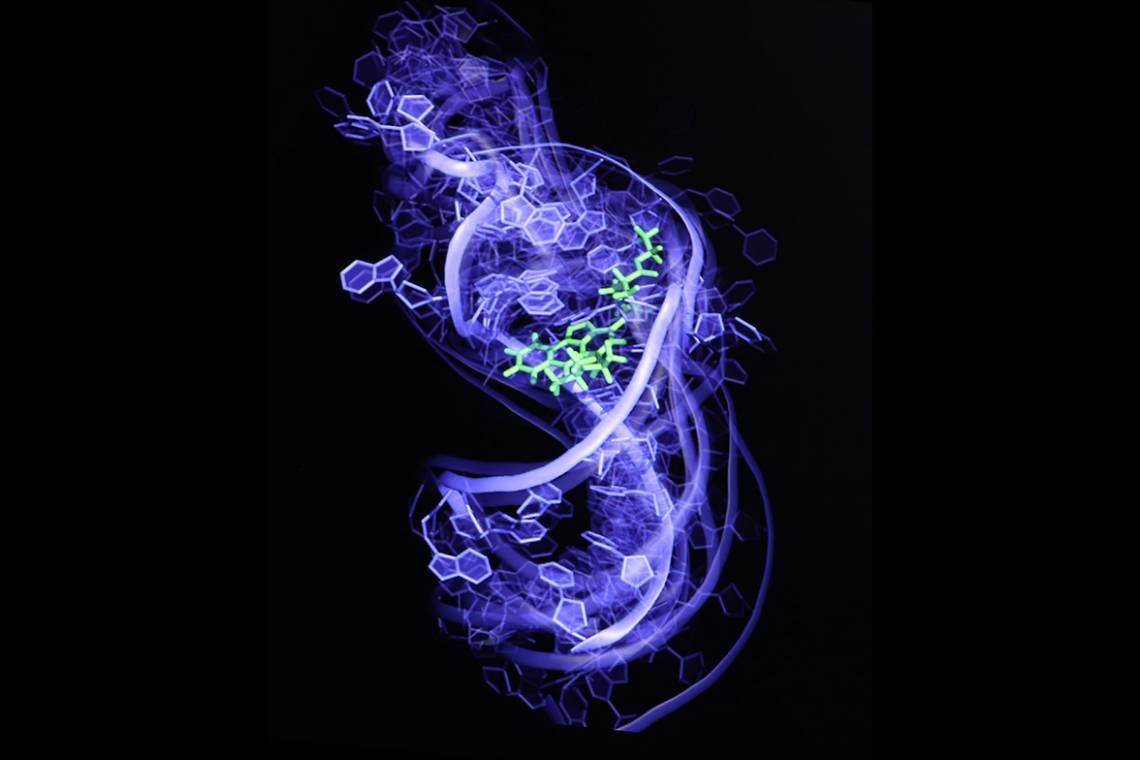
Al-Hashimi and his team have invented a technique that can capture the many states of an RNA molecule and screen hundreds of thousands or perhaps even millions of potential drug candidates. In research published May 4 in the journal Nature Structural and Molecular Biology, they show that their technique can pick compounds with anti-HIV activity out of a line-up of 100,000 that do not.
“This could present a new paradigm for drug discovery,” says Al-Hashimi. “Almost every drug is designed to target proteins. By making it possible to accurately target RNA, we are opening the field up to new and potentially life-saving discoveries.”
The “central dogma” of molecular biology held that genetic information flows from DNA, to RNA, to protein, where the action is. But only about 2 percent of RNAs go on to make proteins. Some research indicates that more than 90 percent of the RNA molecules assembled from the template provided by DNA end up being an RNA as their final state.
But drug discovery efforts over the last fifty years have overlooked nearly all these “non-coding” RNAs, Al-Hashimi said. One clear reason for this glaring omission is the fact that RNA is one of the most flexible, dynamic molecules around. It doesn’t have the typical nooks and crannies that drug developers use to target proteins, and even if it did, a given RNA probably wouldn’t sit still long enough for a scientist to capture it on film.
“This is a long-standing problem,” says Al-Hashimi. “The motion of life is in these molecules. But nobody can predict which drugs will bind RNA, in large part because we don’t have good movies of them.”
Most methods that guide the discovery of drugs either rely on a still image captured in the laboratory that doesn’t show the molecule in action, or movies generated on a computer that are based on calculations, not real data.
In 2011, Al-Hashimi combined the two, harnessing both nuclear magnetic resonance imaging and computationally generated movies to create the first movie of an RNA molecule -- in this case from the virus HIV -- as it danced from one shape to another. His group then took individual frames of the movie, each depicting a different shape of the RNA, and ran them through a computer program to identify molecules that bind to the RNA.
“It was a proof of principle that you can conquer the flexibility of RNA,” says Al-Hashimi. “That was promising, but it wasn’t a rigorous test. A more rigorous test is whether you can fish the needle from the haystack.”
In the present study, Al-Hashimi and his team took 78 compounds known to bind that same RNA target from HIV and added them to a chemical library of 100,000 compounds that they had shown incapable of binding. When they used their technique to screen all the compounds, they were able to pull out the 78 with known anti-HIV activity.
“The key in this study is that we know whether the drugs bind or not, which gives us a means to evaluate how accurate the shapes of our RNA are,” said lead study author Laura Ganser, a graduate student in Al-Hashimi’s lab. “Since it performed so well, we feel that the shapes are accurate, so now we can find and design new drugs.”
They also showed that if they used poor-quality movies, the quality of their predictions went down. The more accurate the data, the more accurate the predictions.
“You can always get the right answer for the wrong reasons,” Ganser said. “By doing this particular exercise and varying the amount of data that goes into the movie, we were convinced this was real.”
The researchers showed that the technique could predict not only what molecules would bind the RNA, but also which particular shape -- linear, L-shaped, S-shaped -- they preferred. That finding is important because unlike small molecules that target proteins by competing for a particular binding site, compounds that target RNA work by locking the molecule in an inactive conformation so it can’t function properly.
Al-Hashimi and his team are currently using the technique to screen millions of compounds for anti-HIV activity before building smaller libraries for further testing.
The research was supported by the National Institutes of Health (P50 GM103297, R01 AI066975, T32 GM08487, and F31 GM119306).
Disclosure - Hashim Al-Hashimi is an advisor to and holds an ownership interest in Nymirum Inc, an RNA-based drug discovery company. Some of the technology used in this paper has been licensed to Nymirum.
CITATION: "High performance virtual screening by targeting a high-resolution RNA dynamic ensemble," Laura R. Ganser, Janghyun Lee, Atul Rangadurai, Dawn K. Merriman, Megan L. Kelly, Aman D. Kansal, Bharathwaj Sathyamoorthy, and Hashim M. Al-Hashimi. Nature Structural and Molecular Biology, May 4, 2018.
DOI: 10.1038/s41594-018-0062-4