DURHAM, N.C. – Tumors are notoriously mixed up; cells from one part often express different genes and adopt different sizes and shapes than cells from another part of that same tumor.
Still, a team of researchers were surprised when they recently spotted a miniature stomach, duodenum, and small intestine hidden among the cells of lung tumor samples.
They discovered that these cells had lost a gene called NKX2-1 that acts as a master switch, flipping a network of genes to set the course for a lung cell. Without it, the cells follow the path of their nearest developmental neighbor -- the gut -- much like a train jumping tracks when a railroad switch fails.
The findings, published March 26 in the journal Developmental Cell, underscore the amazing resilience and plasticity of cancer cells. Such plasticity can presumably enable tumors to develop drug resistance, arguably the biggest challenge to successful cancer treatment.
“Cancer cells will do whatever it takes to survive,” said Purushothama Rao Tata, Ph.D., lead study author and assistant professor of cell biology at Duke University School of Medicine and a member of the Duke Cancer Institute. “Upon treatment with chemotherapy, lung cells shut down some of the key cell regulators and pick up the characteristics of other cells in order to gain resistance.”
Tata has spent most of his career studying the cell types that make up normal lung tissue and how these cells display flexibility during regeneration following an injury. Tata began to wonder whether some of the same rules that he had found governing the normal development and regeneration of tissues might also be responsible for the jumbled nature of tumor cells.
He decided to focus on non-small cell lung cancer, which accounts for 80 to 85 percent of all lung cancer cases. Lung cancer is the leading cause of cancer deaths worldwide, and has one of the lowest survival rates among all cancers. Tata analyzed data from the Cancer Genome Atlas Research Network, a large consortium that has profiled the genomes of thousands of samples from 33 different types of cancer. He found that a large proportion of non-small cell lung cancer tumors lacked NKX2-1, a gene known to specify the lung lineage. Instead, many of them expressed a number of genes associated with esophagus and gastrointestinal organs.
In the absence of NKX2-1, Tata hypothesized, lung tumor cells would lose their lung identity and take on the characteristics of other cells. Because during development lung cells and gut cells are derived from the same parent, or progenitor, cells, it made sense that once the lung cells lost their way they would follow the path of their nearest developmental sibling.
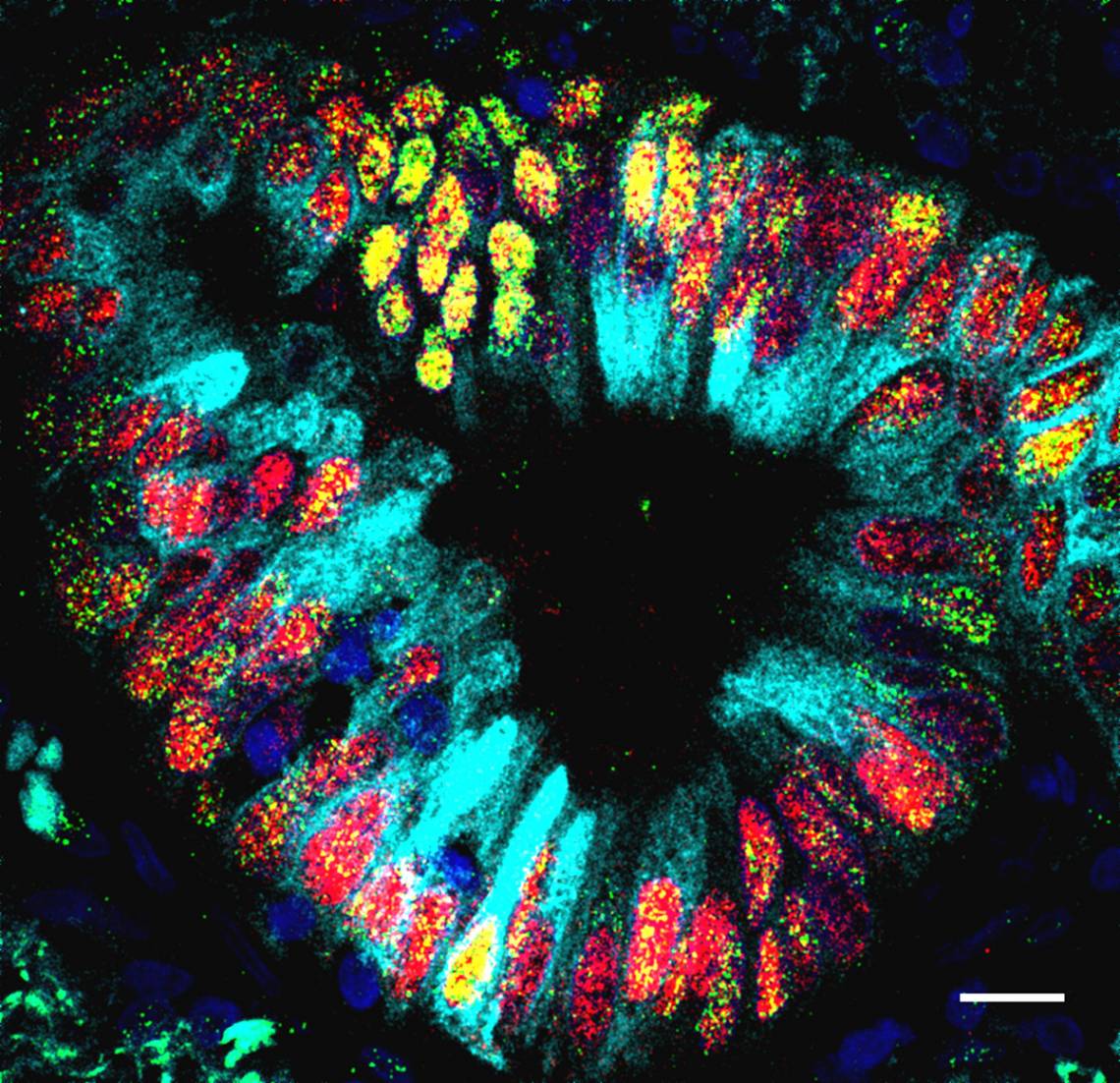
To test whether this was the case, Tata and his colleagues generated different mouse models. First, they knocked out the NKX2-1 gene in the lung tissue of mice. Under the microscope, they noticed features that normally only appear in the gut, such as crypt-like structures and gastric tissues. Amazingly, these structures produced digestive enzymes, as if they resided in the stomach and not the lung.
Having shown that a simple genetic tweak could prompt lung cells to switch developmental tracks, Tata wondered if another tweak or two could fuel them to form tumors. This time, in addition to knocking out NKX2-1, they activated the oncogenes SOX2 or KRAS. The team found that mice with the superimposed SOX2 mutations developed tumors that looked as if they belonged in the foregut; those with KRAS mutations developed tumors that resembled parts of the mid- and hindgut.
Tata and colleagues then wanted to know if these genes were sufficient to alter the fate of lung cells, or if they needed additional signals from their native microenvironment. To answer this question, they developed a novel “mini-lung tumoroid” system -- miniaturized versions of lung tumor tissue -- and found that manipulation of genetics was enough for the lung cells to show such plasticity.
“Cancer biologists have long suspected that cancer cells could shape shift in order to evade chemotherapy and acquire resistance, but they didn’t know the mechanisms behind such plasticity,” said Tata. “Now that we know what we are dealing with in these tumors – we can think ahead to the possible paths these cells might take and design therapies to block them.”
In the future, Tata plans to use his mini-lung tumoroid system to further explore the mechanisms of resistance in lung cancer cells.
This research was supported by the New York Stem Cell Foundation, the National Institutes of Health/National Heart, Lung, and Blood Institute Early Career Research New Faculty (P30) award (5P30HL101287-02) and RO1 (RO1HL118185), the Harvard Ludwig Cancer Center, the Massachusetts Eye and Ear Infirmary, a Harvard Stem Cell Institute Junior Investigator Grant, an NIH MSTP training grant (T32GM007205), the Medical Scientist Training Program (GM007101), an NIH/NHLBI Career Development Award (K99HL127181), the Whitehead Scholar Program, the Maroni Research Scholar Program, and the HHMI Faculty Scholar Program.
CITATION: "Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity," Purushothama Rao Tata, Ryan Dz-Wei Chow, Srinivas Vinod Saladi, Aleksandra Tata, Arvind Konkimalla, Anne Bara, Daniel Montoro, Lida P. Hariri, Angela R. Shih, Mari Mino-Kenudson, Hongmei Mou, Sioko Kimura, Leif W. Ellisen, Jayaraj Rajagopal. Developmental Cell, March 26, 2018. DOI: 10.1016/j.devcel.2018.02.024