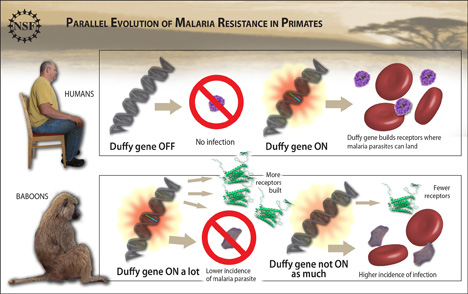
Evolutionarily speaking, baboons may be our more distant cousins among primates. But when it comes to our experiences with malaria over the course of time, it seems the stories of our two species have followed very similar plots.
In humans, subtle variation in one particular gene that controls whether a protein on the surface of red blood cells gets made or not literally spells the difference between susceptibility or resistance to one form of malaria. That's because the blood protein serves as the entry point for Plasmodium vivax, one of several malaria-causing parasites that infect humans.
Now, researchers at the Duke Institute for Genome Sciences & Policy report that variation in precisely the same regulatory gene also influences baboons' chances of getting sick, by ratcheting their susceptibility to another, closely related parasite up or down.
"It's a nice example of how -- in the vastness of the genome -- the same gene was modified in the same way in two different species to produce the same kind of resistance," says Greg Wray, director of the IGSP's Center for Evolutionary Genomics. "That's a pretty remarkable thing when you think of all the different ways malaria resistance might have evolved."
The findings, which appeared online in Nature on June 24, also mark a turning point in primate research: they are the first to connect any functionally important genetic variation in wild primates to complex, real-life consequences for the animals.
The yellow baboons in question live in Kenya's Amboseli National Park and have been the subject of ongoing observation for nearly 40 years, making them one of the best-studied wild mammal populations in the world from a behavioral and life history standpoint.
"It used to be that our work was limited to ‘skin-out' biology," says Susan Alberts, an associate professor of biology and IGSP member who has been recording the habits of the baboons for the last 25 years. Today, thanks to a growing library of sequenced primate genomes including our own, scientists can begin to delve deeper.
Graduate student Jenny Tung spent three summers out on the East African savanna, watching the baboons, collecting their DNA-laden feces, and with the help of an expert team of Kenyan field assistants, very carefully drawing blood from darted animals. Successfully darting baboons is no small feat, Tung said. You have to be within meters of the animal you are targeting, and at the same time make sure that none of the baboons catch you in the act. If they did, it would send the troop running and screaming and, in technical terms, "really mess up the field data." In the evenings, Tung processed and stored her hard-won samples in a makeshift refrigerator before shipping them off to Duke.
Once back at the lab, Tung found something in those blood samples that came as a surprise despite all the years of study. More than half of the Amboseli baboons -- some 60 percent -- were infected with the malaria-like parasite known as Hepatocystis.
"We had no idea so many of them were carrying this parasite," Alberts says. For years, researchers have tracked the baboons for any signs of injury or illness. But although the infection probably compromises the animals, they don't develop cyclical fever spikes or other immediately obvious symptoms like humans with malaria do.
In search of a genetic basis for differences in the baboons' vulnerability to infection, the researchers zeroed in on the DNA sequence surrounding the DARC gene, the same region that has been traced to malaria protection among people. Although the specifics differ from those in humans, they found that a single letter change to the genetic code -- a switch from an A to a G -- lends some baboons the ability to better fend off infection. In fact, they show, one G is good, but two are even better.
Further analysis of the baboons' blood and in cell culture confirmed that the variants influence infection rates through changes in the activity of the DARC gene. Comparison of the Amboseli baboon sequences to two other populations also showed that the DNA sequence has undergone a relatively rapid rate of evolutionary change, the mark of natural selection for malaria resistance.
The newfound parallels between baboons and humans bring the long history of conflict between parasite and host into high relief. "It's a struggle out there," Alberts says. "We often think of malaria as a contemporary problem, but it's a deep part of our history."
The study also shows the power of coupling genomics with dedicated fieldwork. "Part of what we want to do is push the envelope and show that this is doable," Wray says. With the proof of principle in hand, the next big challenge is to begin to unravel the genomic differences that may be responsible for fuzzier behavioral traits, such as social status or aggression, he added.
"It's getting easier and easier to generate genetic data," Tung says. "But it's never going to be easy to have long-term field data -- especially for primates. It takes years and years before you see the fruit of those labors. We're just at the point where it's going to really start paying off."
Collaborators on the study include Alexander Primus, Andrew Bouley and Tonya Severson, all of Duke. The work was funded by the National Science Foundation, the American Society of Primatologists, Duke University, the Duke chapter of Sigma Xi, and the Duke Institute for Genome Sciences & Policy.