
Mysterious Family of Microbial Proteins Hijack Crops’ Cellular Plumbing
Duke researchers may have come up with a way to disarm them, and thus help prevent $220 billion in annual crop damage
Ever since their discovery in the early 1990s, this family of proteins has been of great interest to those who study plant disease. They are key weapons in the bacterial arsenal; knocking them out in a lab renders otherwise-dangerous bacteria harmless. But, despite decades of effort, many questions about how they work remain unanswered.
Researchers had identified a number of proteins in the AvrE/DspE family that suppressed the plant’s immune system, or that caused dark water-soaked spots on a plant’s leaves -- the first telltale signs of infection. They even knew the underlying sequence of amino acids that linked to form the proteins, like beads on a string. But they didn’t know how this string of amino acids folded into a 3D shape, so they couldn’t easily explain how they worked.
Part of the problem is that the proteins in this family are huge. Whereas an average bacterial protein might be 300 amino acids long; AvrE/DspE-family proteins are 2000.
Researchers have looked for other proteins with similar sequences for clues, but none with any known functions showed up.
“They’re weird proteins,” He said.
So they turned to a computer program released in 2021 called AlphaFold2, which uses artificial intelligence to predict what 3D shape a given string of amino acids will take.
The researchers knew that some members of this family help the bacteria evade the plant’s immune system. But their first glimpse of the proteins’ 3D structure suggested an additional role.
“When we first saw the model, it was nothing like what we had thought,” said study co-author Pei Zhou, a professor of biochemistry at Duke whose lab contributed to the findings.
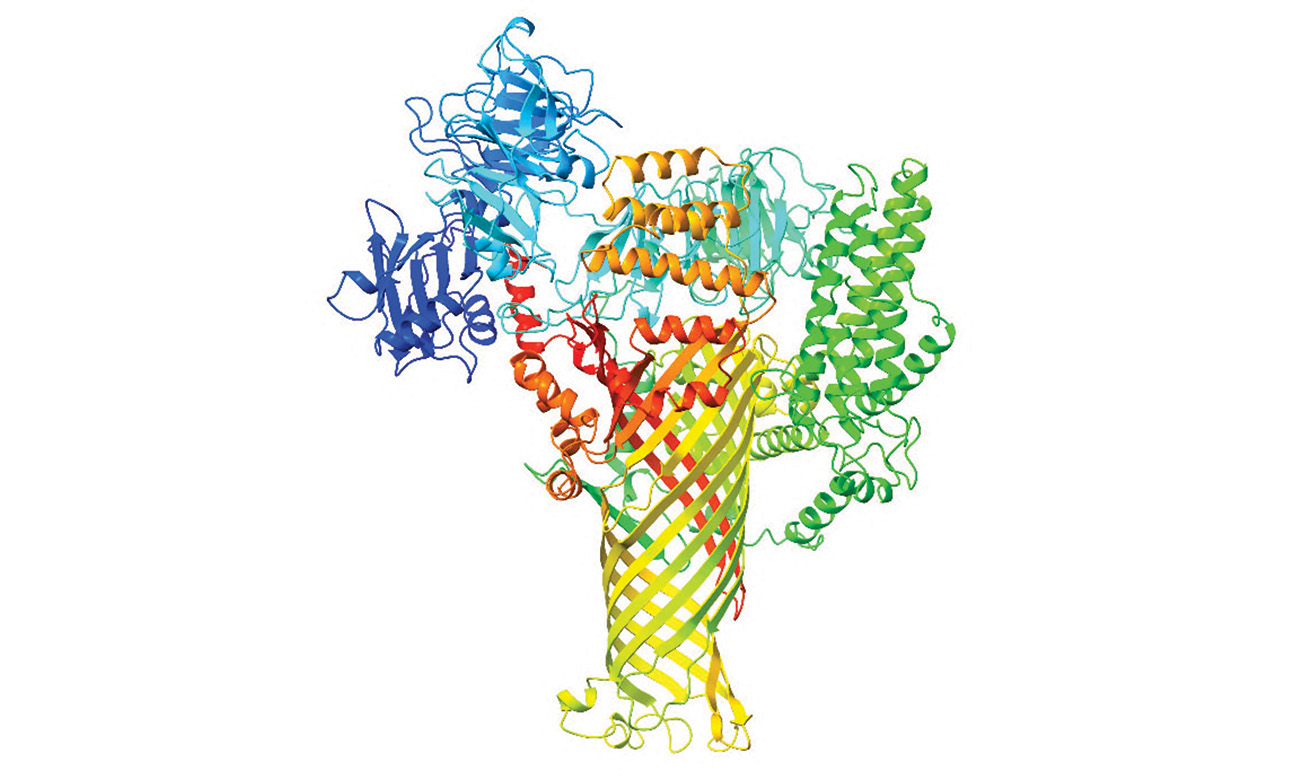
The researchers looked at AI predictions for bacterial proteins that infect crops including pears, apples, tomatoes and corn, and they all pointed to a similar 3D structure. They appeared to fold into a tiny mushroom with a cylindrical stem, like a straw.
The predicted shape matched up well with images of a bacterial protein that causes fire blight disease in fruit trees that was captured using a cryo-electron microscope. From the top down, this protein looked very much like a hollow tube.
Which got the researchers thinking: Perhaps bacteria use these proteins to punch a hole in the plant cell membrane, to "force the host for a drink" during infection, He said.
Once bacteria enter the leaves, one of the first areas they come across is a space between cells called the apoplast. Normally, plants keep this area dry to enable gas exchange for photosynthesis. But when bacteria invade, the inside of the leaf becomes waterlogged, creating a moist cozy haven for them to feed and multiply.
Further examination of the predicted 3D model for the fire blight protein revealed that, while the outside of the straw-like structure is water-resistant, its hollow inner core has a special affinity for water.
To test the water channel hypothesis, the team joined forces with Duke biology professor Ke Dong and co-first-author Felipe Andreazza, a postdoctoral associate in her lab. They added the gene readouts for the bacterial proteins AvrE and DspE to frog eggs, using the eggs as cellular factories for making the proteins. The eggs, placed in a dilute saline solution, quickly swelled and burst with too much water.
The researchers also tried to see if they could disarm these bacterial proteins by blocking their channels. Nomura focused on a class of tiny spherical nanoparticles called PAMAM dendrimers. Used for more than two decades in drug delivery, these dendrimers can be made with precise diameters in a lab.
“We were tinkering with the hypothesis that if we found the right diameter chemical, maybe we could block the pore,” He said.
After testing different sized particles, they identified one they thought might be just the right size for jamming the water channel protein produced by the fire blight pathogen, Erwinia amylovora.
They took frog eggs engineered to synthesize this protein and doused them with the PAMAM nanoparticles, and water stopped flowing into the eggs. They didn’t swell.
They also treated Arabidopsis plants infected with the pathogen Pseudomonas syringae, which causes bacterial speck. The channel-blocking nanoparticles prevented the bacteria from taking hold, reducing pathogen concentrations in the plants’ leaves by 100-fold.
The compounds were effective against other bacterial infections too. The researchers did the same thing with pear fruits exposed to the bacteria that cause fire blight disease, and the fruits never developed symptoms -- the bacteria didn’t make them sick.
“It was a long shot, but it worked,” He said. “We're excited about this.”
The findings could offer a new line of attack against many plant diseases, the researchers said.
Plants produce 80% of the food we eat. And yet more than 10% of global food production -- crops such as wheat, rice, maize, potato and soybean -- are lost to plant pathogens each year, costing the global economy a whopping $220 billion.
The team has filed a provisional patent on the approach.
The next step, said Zhou and co-first-author Jie Cheng, a Ph.D. student in Zhou’s lab, is to figure out how this protection works, by getting a more detailed look at how the channel-blocking nanoparticles and the channel proteins interact.
“If we can image those structures we can have a better understanding and come up with better designs for crop protection,” Zhou said.
This work was supported by the National Institute of Allergy and Infectious Diseases (1R01AI155441) and the National Institute of General Medical Sciences (GM145026), both at the National Institutes of Health, Duke Science and Technology, and the Howard Hughes Medical Institute.
Citation
"Bacterial Pathogens Deliver Water/Solute-Permeable Channels as a Virulence Strategy," Kinya Nomura, Felipe Andreazza, Jie Cheng, Ke Dong, Pei Zhou, and Sheng Yang He. Nature, Sept. 13, 2023. DOI: 10.1038/s41586-023-06531-5